Find Batch-biased Features in SVGs
Christine Hou
Department of Biostatistics, Johns Hopkins Universitychris2018hou@gmail.com Source:
vignettes/spe.Rmd
spe.RmdIntroduction
BatchSVG is an R/Bioconductor package for spatial
transcriptomics data quality control (QC). As a feature-based QC method,
the package provides functions to identify biased features associated
with batch effect(s) (e.g. sample, slide, and sex) in spatially variable
genes (SVGs) using the binomial deviance model, aiming to improve the
downstream clustering performance and remove technical noise caused by
batch effects. The package works with SpatialExperiment
objects.
Installation
if (!requireNamespace("BiocManager")) {
install.packages("BiocManager")
}
BiocManager::install("BatchSVG")Install the development version from GitHub.
remotes::install("christinehou11/BatchSVG")Biased Feature Identification
In this section, we will include the standard workflow for using
BatchSVG to show how the method helps to detect and
visualize biased features in SVGs.
Data
We will use the spatially-resolved transcriptomics (SRT) dataset from the spatialLIBD package.
spatialLIBD_spe <- fetch_data(type = "spe")
spatialLIBD_spe
class: SpatialExperiment
dim: 33538 47681
metadata(0):
assays(2): counts logcounts
rownames(33538): ENSG00000243485 ENSG00000237613 ... ENSG00000277475
ENSG00000268674
rowData names(9): source type ... gene_search is_top_hvg
colnames(47681): AAACAACGAATAGTTC-1 AAACAAGTATCTCCCA-1 ...
TTGTTTCCATACAACT-1 TTGTTTGTGTAAATTC-1
colData names(69): sample_id Cluster ... array_row array_col
reducedDimNames(6): PCA TSNE_perplexity50 ... TSNE_perplexity80
UMAP_neighbors15
mainExpName: NULL
altExpNames(0):
spatialCoords names(2) : pxl_col_in_fullres pxl_row_in_fullres
imgData names(4): sample_id image_id data scaleFactorWe will use the spatially variable genes set previously generated. The result is generated from nnSVG package.
libd_nnsvgs <- read.csv(
system.file("extdata","libd-all_nnSVG_p-05-features-df.csv",
package = "BatchSVG"),
row.names = 1, check.names = FALSE)Perform Feature Selection using featureSelect()
We will perform feature selection on a subset of spatial transcriptomics data (input) using a predefined set of spatially variable genes (SVGs). Specifically, we will compute the number of standard deviations for the relative change in deviance (nSD_dev_{batch effect}) and rank difference (nSD_rank_{batch effect}) before and after adjusting for batch effects.
The featureSelect() function enables feature selection
while accounting for multiple batch effects. It returns a
list of data frames, where each batch effect is
associated with a corresponding data frame containing key results,
including:
Relative change in deviance before and after batch effect adjustment
Rank differences between the batch-corrected and uncorrected results
Number of standard deviations (nSD) for both relative change in deviance and rank difference
We will use the example of applying featureSelect() to
the sample dataset while adjusting for the batch effect of
subject.
list_batch_df <- featureSelect(input = spatialLIBD_spe,
batch_effect = "subject", VGs = libd_nnsvgs$gene_id)
Running feature selection with batch...
Batch Effect: subject
Running feature selection without batch...
Calculating deviance and rank difference...To suppress the message, let verbose = FALSE.
list_batch_df <- featureSelect(input = spatialLIBD_spe,
batch_effect = "subject", VGs = libd_nnsvgs$gene_id, verbose = FALSE)head(list_batch_df$subject)
gene_id gene_name dev_default rank_default dev_subject rank_subject
1 ENSG00000187608 ISG15 43591.43 1151 43304.39 1157
2 ENSG00000131584 ACAP3 39115.97 1498 38826.05 1509
3 ENSG00000242485 MRPL20 47074.23 920 46912.53 918
4 ENSG00000160075 SSU72 47815.13 862 47742.10 852
5 ENSG00000078369 GNB1 54464.75 477 54385.81 462
6 ENSG00000187730 GABRD 51521.13 646 51099.35 648
d_diff nSD_dev_subject r_diff nSD_rank_subject
1 0.006628419 -0.04505973 6 0.15937488
2 0.007467317 -0.03697398 11 0.29218728
3 0.003446929 -0.07572463 -2 -0.05312496
4 0.001529688 -0.09420402 -10 -0.26562480
5 0.001451448 -0.09495814 -15 -0.39843720
6 0.008254157 -0.02939000 2 0.05312496Visualize SVG Selection Using svg_nSD for Batch
Effects
The svg_nSD() function generates visualizations to
assess batch effects in spatially variable genes (SVGs). It produces bar
charts showing the distribution of SVGs based on relative change in
deviance and rank difference, with colors representing different nSD
intervals. Additionally, scatter plots compare deviance and rank values
with and without batch effects.
By interpreting these plots, we can determine appropriate nSD thresholds for filtering biased features. The left panels illustrate the distribution of SVGs in terms of deviance and rank difference, while the right panels compare values before and after accounting for batch effects.
plots <- svg_nSD(list_batch_df = list_batch_df,
sd_interval_dev = 3, sd_interval_rank = 3)Figure 1. Visualizations of nSD_dev and nSD_rank threshold selection
plots$subject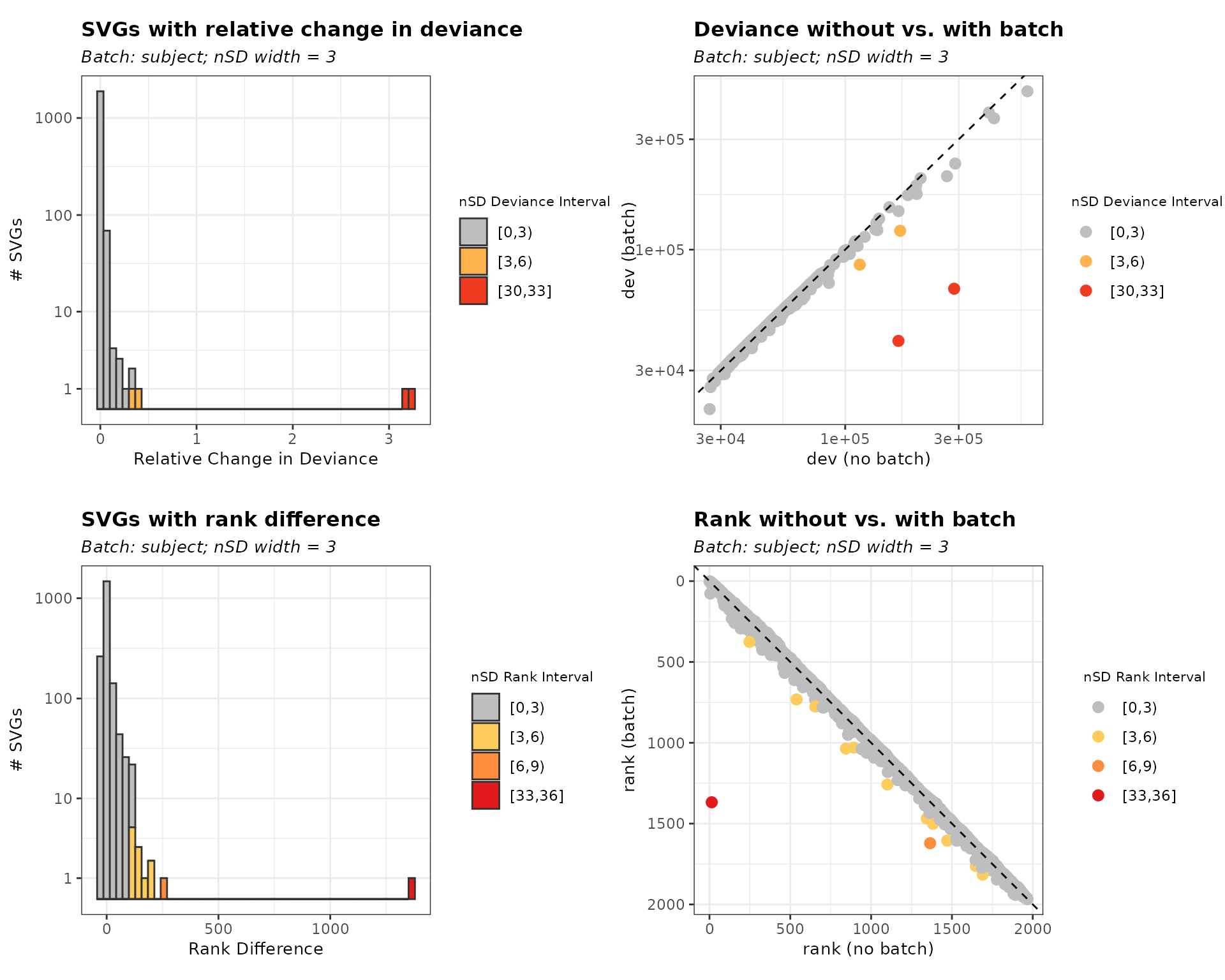
Identify Biased Genes Using biasDetect()
The function biasDetect() is designed to identify and
filter out biased genes across different batch effects. Using threshold
values selected from the visualization results generated by
svg_nSD(), this function systematically detects outliers
that exceed a specified number of standard deviation (nSD) threshold in
either relative deviance change, rank difference, or both.
The function outputs visualizations comparing deviance and rank values with and without batch effects. Genes with high deviations, highlighted in color, are identified as potentially biased and can be excluded based on the selected nSD thresholds.
The function offers flexibility in customizing the plot aesthetics, allowing users to adjust the data point size (plot_point_size), shape (plot_point_shape), annotated text size (plot_text_size), and data point color palette (plot_palette). Default values are provided for these parameters if not specified. Users should refer to ggplot2 aesthetic guidelines to ensure appropriate values are assigned for each parameter.
We will use nSD_dev = 3 and nSD_rank = 3 as
the example. The user should adjust the value based on their dataset
features.
Usage of Different Threshold Options
-
threshold = "dev": Filters biased genes based only on the relative change in deviance. Genes with deviance changes exceeding the specifiednSD_devthreshold are identified as batch-affected and can be removed.
bias_dev <- biasDetect(list_batch_df = list_batch_df,
threshold = "dev", nSD_dev = 3)Table 1. Outlier Genes defined by nSD_dev only
head(bias_dev$subject$Table)
gene_id gene_name dev_default rank_default dev_subject rank_subject
1 ENSG00000255823 MTRNR2L8 286927.9 5 67738.95 77
2 ENSG00000087250 MT3 115108.2 21 86093.50 31
3 ENSG00000256618 MTRNR2L1 167381.9 14 40299.65 1368
4 ENSG00000198840 MT-ND3 170192.2 12 120647.03 17
d_diff nSD_dev_subject r_diff nSD_rank_subject nSD_bin_dev dev_outlier
1 3.2357886 31.079310 72 1.9124986 [30,33] TRUE
2 0.3370142 3.139375 10 0.2656248 [3,6) TRUE
3 3.1534316 30.285509 1354 35.9655982 [30,33] TRUE
4 0.4106625 3.849237 5 0.1328124 [3,6) TRUEWe can change the data point size using plot_point_size.
# size default = 3
bias_dev_size <- biasDetect(list_batch_df = list_batch_df,
threshold = "dev", nSD_dev = 3, plot_point_size = 4)Figure 2. Customize point size
plot_grid(bias_dev$subject$Plot, bias_dev_size$subject$Plot)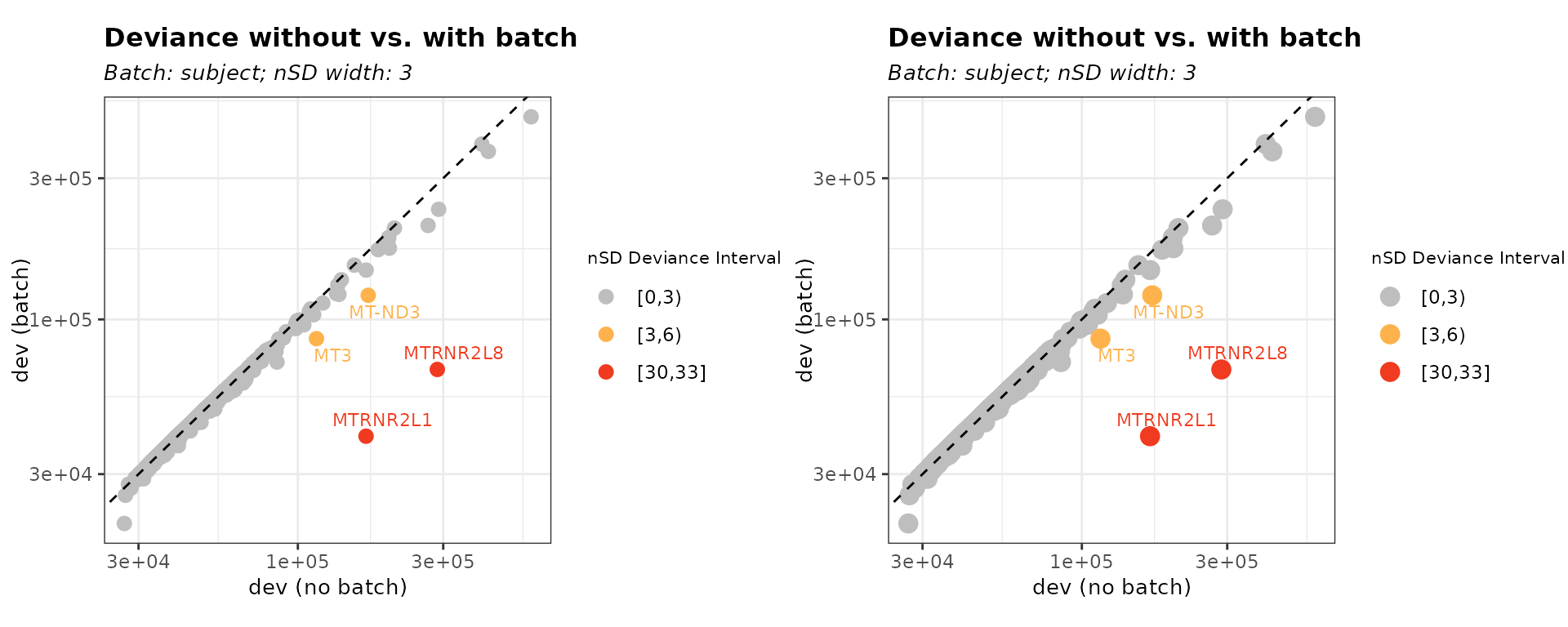
-
threshold = "rank": Identifies biased genes based solely on rank difference. Genes with rank shifts exceedingnSD_rankare considered biased.
bias_rank <- biasDetect(list_batch_df = list_batch_df,
threshold = "rank", nSD_rank = 3)Table 2. Outlier Genes defined by nSD_rank only
head(bias_rank$subject$Table)
gene_id gene_name dev_default rank_default dev_subject rank_subject
1 ENSG00000184144 CNTN2 44388.43 1101 42016.77 1258
2 ENSG00000136541 ERMN 48106.50 844 44879.30 1036
3 ENSG00000013297 CLDN11 53385.84 539 49776.21 732
4 ENSG00000164124 TMEM144 39407.07 1471 37587.88 1605
5 ENSG00000204655 MOG 37293.40 1648 35613.38 1762
6 ENSG00000091129 NRCAM 51388.42 655 49006.18 776
d_diff nSD_dev_subject r_diff nSD_rank_subject nSD_bin_rank rank_outlier
1 0.05644564 0.4351052 157 4.170309 [3,6) TRUE
2 0.07190853 0.5841448 192 5.099996 [3,6) TRUE
3 0.07251725 0.5900120 193 5.126559 [3,6) TRUE
4 0.04839817 0.3575395 134 3.559372 [3,6) TRUE
5 0.04717397 0.3457399 114 3.028123 [3,6) TRUE
6 0.04861096 0.3595904 121 3.214060 [3,6) TRUEWe can change the data point shape using plot_point_shape.
Figure 3. Customize point shape
# shape default = 16
bias_rank_shape <- biasDetect(list_batch_df = list_batch_df,
threshold = "rank", nSD_rank = 3, plot_point_shape = 2)
plot_grid(bias_rank$subject$Plot, bias_rank_shape$subject$Plot)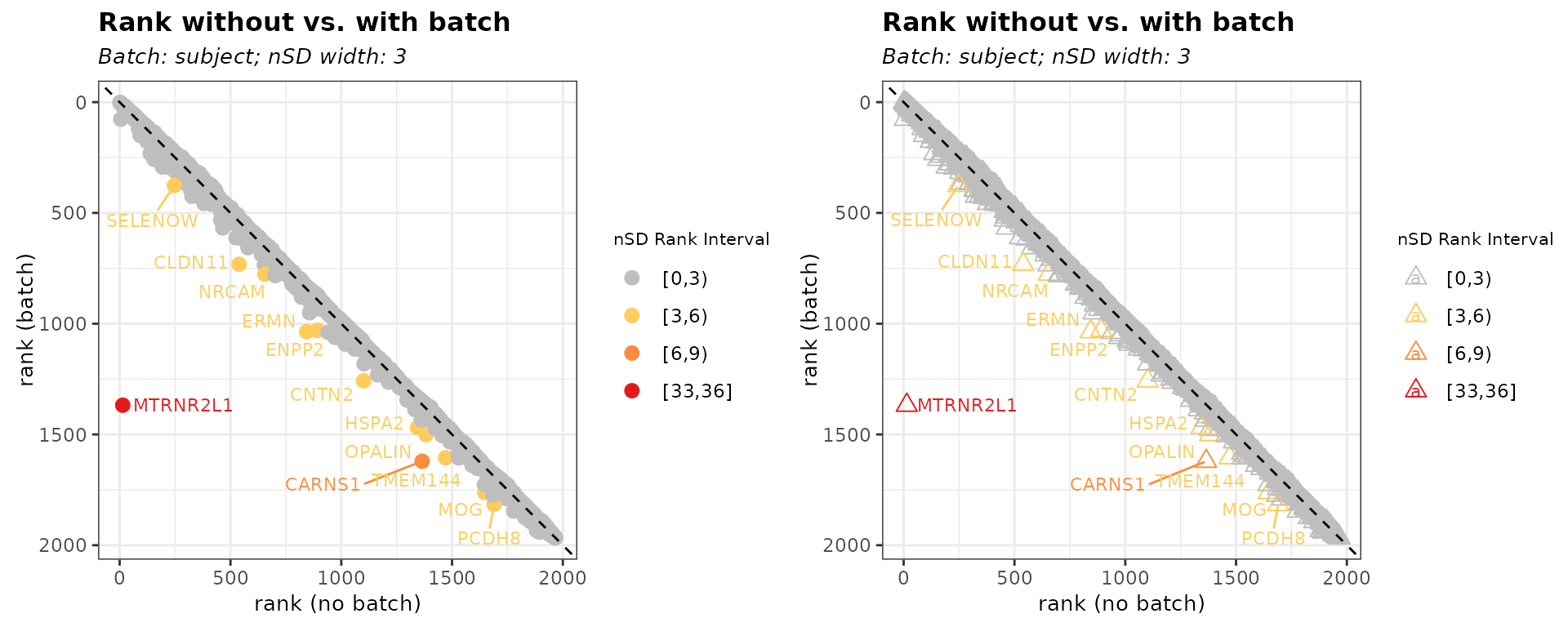
-
threshold = "both": Detects biased genes based on both deviance change and rank difference, providing a more stringent filtering approach.
bias_both <- biasDetect(list_batch_df = list_batch_df, threshold = "both",
nSD_dev = 3, nSD_rank = 3)Table 3. Outlier Genes defined by nSD_dev and nSD_rank
head(bias_both$subject$Table)
gene_id gene_name dev_default rank_default dev_subject rank_subject
1 ENSG00000184144 CNTN2 44388.43 1101 42016.77 1258
2 ENSG00000136541 ERMN 48106.50 844 44879.30 1036
3 ENSG00000013297 CLDN11 53385.84 539 49776.21 732
4 ENSG00000164124 TMEM144 39407.07 1471 37587.88 1605
5 ENSG00000204655 MOG 37293.40 1648 35613.38 1762
6 ENSG00000091129 NRCAM 51388.42 655 49006.18 776
d_diff nSD_dev_subject r_diff nSD_rank_subject nSD_bin_dev dev_outlier
1 0.05644564 0.4351052 157 4.170309 [0,3) FALSE
2 0.07190853 0.5841448 192 5.099996 [0,3) FALSE
3 0.07251725 0.5900120 193 5.126559 [0,3) FALSE
4 0.04839817 0.3575395 134 3.559372 [0,3) FALSE
5 0.04717397 0.3457399 114 3.028123 [0,3) FALSE
6 0.04861096 0.3595904 121 3.214060 [0,3) FALSE
nSD_bin_rank rank_outlier
1 [3,6) TRUE
2 [3,6) TRUE
3 [3,6) TRUE
4 [3,6) TRUE
5 [3,6) TRUE
6 [3,6) TRUEWe can change the data point color using
plot_palette. The color palette here
can be referenced on since the function uses RColorBrewer
to generate colors.
Figure 4. Customize the palette color
# color default = "YlOrRd"
bias_both_color <- biasDetect(list_batch_df = list_batch_df,
threshold = "both", nSD_dev = 3, nSD_rank = 3, plot_palette = "Greens")
plot_grid(bias_both$subject$Plot, bias_both_color$subject$Plot,nrow = 2)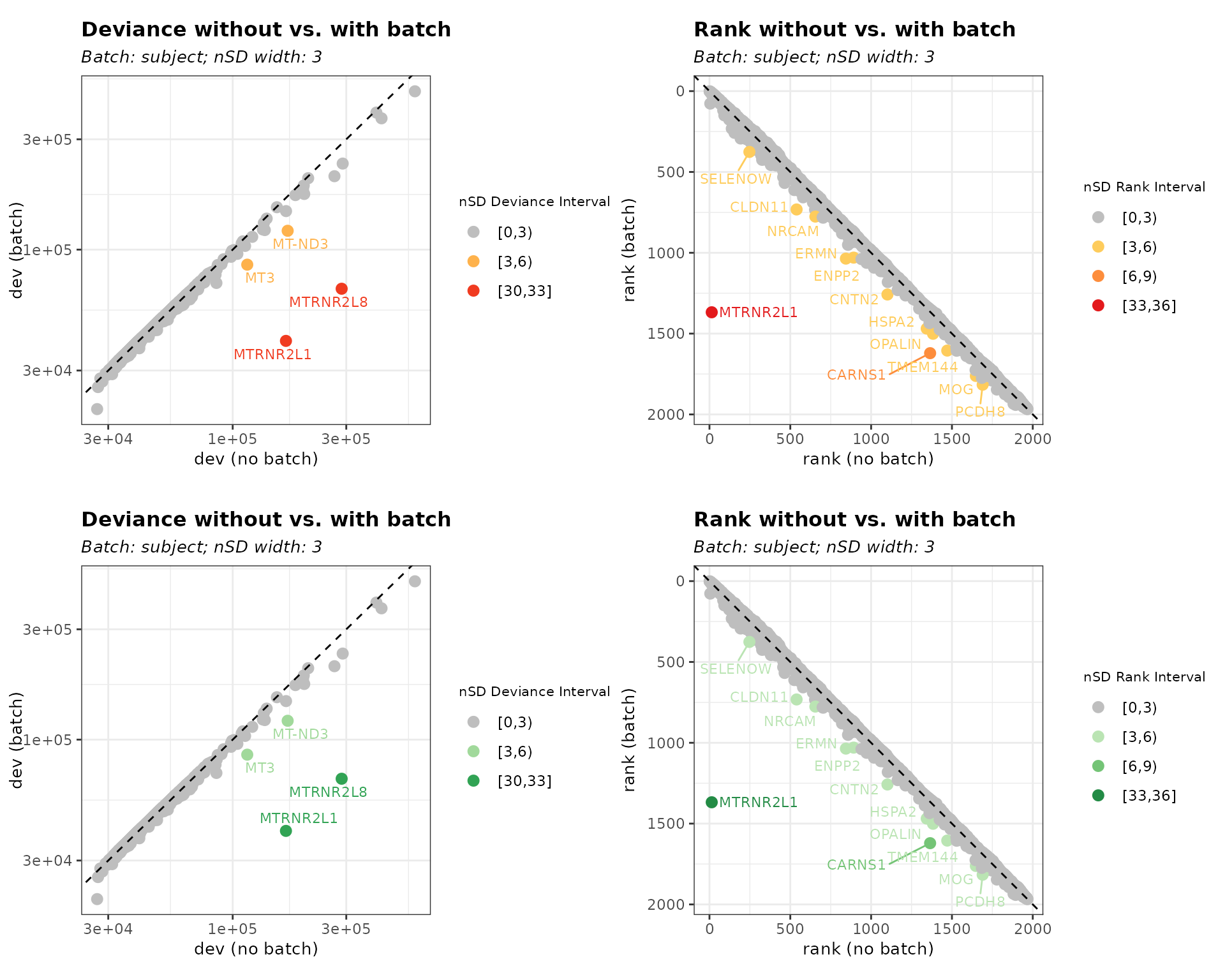
We can change the text size using plot_text_size. We also specify the color palettes for both batch effects at the same time.
Figure 5. Customize text size and color palette
# text size default = 3
bias_both_color_text <- biasDetect(list_batch_df = list_batch_df,
threshold = "both", nSD_dev = 3, nSD_rank = 3,
plot_palette = c("Blues"), plot_text_size = 4)
plot_grid(bias_both$subject$Plot, bias_both_color_text$subject$Plot,nrow = 2)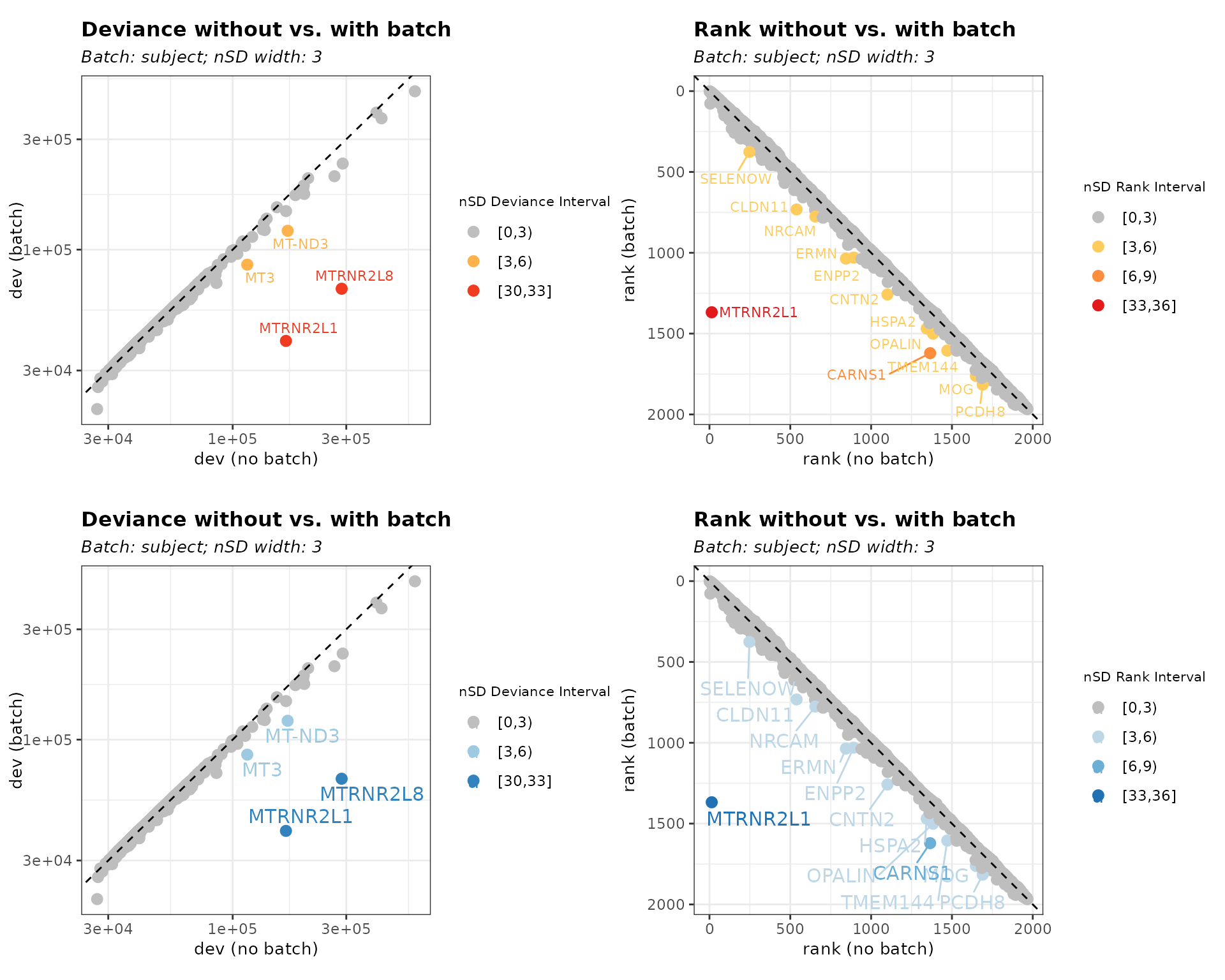
Refine SVGs by Removing Batch-Affected Outliers
Finally, we obtain a refined set of spatially variable genes (SVGs)
by removing the identified outliers based on user-defined thresholds for
nSD_dev and nSD_rank.
Here, we use the results from bias_both, which applied
threshold = "both" to account for both deviance and rank
differences, with the batch effect set to sample ID.
bias_both_df <- bias_both$subject$Table
svgs_filt <- setdiff(libd_nnsvgs$gene_id, bias_both_df$gene_id)
svgs_filt_spe <- libd_nnsvgs[libd_nnsvgs$gene_id %in% svgs_filt, ]
nrow(svgs_filt_spe)
[1] 1951After obtaining the refined set of SVGs, these genes can be further analyzed using established spatial transcriptomics clustering algorithms to explore tissue layers and spatial organization.
R session information
## Session info
sessionInfo()
#> R version 4.5.1 (2025-06-13)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Ubuntu 24.04.3 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats4 stats graphics grDevices utils datasets methods
#> [8] base
#>
#> other attached packages:
#> [1] cowplot_1.2.0 spatialLIBD_1.20.1
#> [3] SpatialExperiment_1.18.1 SingleCellExperiment_1.30.1
#> [5] SummarizedExperiment_1.38.1 Biobase_2.68.0
#> [7] GenomicRanges_1.60.0 GenomeInfoDb_1.44.3
#> [9] IRanges_2.42.0 S4Vectors_0.46.0
#> [11] BiocGenerics_0.54.0 generics_0.1.4
#> [13] MatrixGenerics_1.20.0 matrixStats_1.5.0
#> [15] BatchSVG_1.1.1 BiocStyle_2.36.0
#>
#> loaded via a namespace (and not attached):
#> [1] RColorBrewer_1.1-3 jsonlite_2.0.0 shape_1.4.6.1
#> [4] magrittr_2.0.4 ggbeeswarm_0.7.2 magick_2.9.0
#> [7] farver_2.1.2 rmarkdown_2.30 BiocIO_1.18.0
#> [10] GlobalOptions_0.1.2 fs_1.6.6 ragg_1.5.0
#> [13] vctrs_0.6.5 Rsamtools_2.24.1 config_0.3.2
#> [16] memoise_2.0.1 RCurl_1.98-1.17 paletteer_1.6.0
#> [19] benchmarkme_1.0.8 htmltools_0.5.8.1 S4Arrays_1.8.1
#> [22] AnnotationHub_3.16.1 curl_7.0.0 BiocNeighbors_2.2.0
#> [25] SparseArray_1.8.1 sass_0.4.10 bslib_0.9.0
#> [28] htmlwidgets_1.6.4 desc_1.4.3 plotly_4.11.0
#> [31] cachem_1.1.0 GenomicAlignments_1.44.0 mime_0.13
#> [34] lifecycle_1.0.4 iterators_1.0.14 pkgconfig_2.0.3
#> [37] rsvd_1.0.5 Matrix_1.7-3 R6_2.6.1
#> [40] fastmap_1.2.0 GenomeInfoDbData_1.2.14 shiny_1.11.1
#> [43] clue_0.3-66 digest_0.6.37 colorspace_2.1-2
#> [46] rematch2_2.1.2 AnnotationDbi_1.70.0 scater_1.36.0
#> [49] irlba_2.3.5.1 ExperimentHub_2.16.1 textshaping_1.0.3
#> [52] RSQLite_2.4.3 beachmat_2.24.0 labeling_0.4.3
#> [55] filelock_1.0.3 httr_1.4.7 abind_1.4-8
#> [58] compiler_4.5.1 withr_3.0.2 attempt_0.3.1
#> [61] bit64_4.6.0-1 doParallel_1.0.17 S7_0.2.0
#> [64] BiocParallel_1.42.2 viridis_0.6.5 DBI_1.2.3
#> [67] sessioninfo_1.2.3 rappdirs_0.3.3 DelayedArray_0.34.1
#> [70] rjson_0.2.23 tools_4.5.1 vipor_0.4.7
#> [73] beeswarm_0.4.0 httpuv_1.6.16 glue_1.8.0
#> [76] restfulr_0.0.16 promises_1.3.3 grid_4.5.1
#> [79] cluster_2.1.8.1 gtable_0.3.6 tidyr_1.3.1
#> [82] data.table_1.17.8 BiocSingular_1.24.0 ScaledMatrix_1.16.0
#> [85] XVector_0.48.0 stringr_1.5.2 ggrepel_0.9.6
#> [88] BiocVersion_3.21.1 foreach_1.5.2 pillar_1.11.1
#> [91] limma_3.64.3 later_1.4.4 circlize_0.4.16
#> [94] benchmarkmeData_1.0.4 dplyr_1.1.4 BiocFileCache_2.16.2
#> [97] lattice_0.22-7 rtracklayer_1.68.0 bit_4.6.0
#> [100] tidyselect_1.2.1 locfit_1.5-9.12 ComplexHeatmap_2.24.1
#> [103] scuttle_1.18.0 Biostrings_2.76.0 knitr_1.50
#> [106] gridExtra_2.3 bookdown_0.45 edgeR_4.6.3
#> [109] xfun_0.53 statmod_1.5.0 DT_0.34.0
#> [112] stringi_1.8.7 UCSC.utils_1.4.0 lazyeval_0.2.2
#> [115] yaml_2.3.10 shinyWidgets_0.9.0 evaluate_1.0.5
#> [118] codetools_0.2-20 tibble_3.3.0 BiocManager_1.30.26
#> [121] cli_3.6.5 xtable_1.8-4 systemfonts_1.3.1
#> [124] jquerylib_0.1.4 golem_0.5.1 Rcpp_1.1.0
#> [127] dbplyr_2.5.1 scry_1.20.0 png_0.1-8
#> [130] XML_3.99-0.19 parallel_4.5.1 pkgdown_2.1.3
#> [133] ggplot2_4.0.0 blob_1.2.4 bitops_1.0-9
#> [136] viridisLite_0.4.2 scales_1.4.0 purrr_1.1.0
#> [139] crayon_1.5.3 GetoptLong_1.0.5 rlang_1.1.6
#> [142] KEGGREST_1.48.1